Natural decomposition approach
(Positions available: This project is currently recruiting PhD and BSc students)
In scientific literature, there are several methods to identify modules in complex networks, but all these methods do not exploit the biological knowledge that we have about some networks. On the contrary, these methods are based on optimization processes that could find suboptimal solutions or rely on non-objective parameters without any biological meaning.
To overcome these limitations, we developed the natural decomposition approach (NDA). The NDA defines criteria to identify systems and system-level elements in a regulatory network, and rules to reveal its functional architecture by controlled decomposition (Figure 1). This biologically motivated approach mathematically derives the architecture and system-level elements from the global structural properties of a given regulatory network. It is based on two biological premises:
- A module is a set of genes cooperating to carry out a particular physiological function, thus conferring different phenotypic traits to the cell.
- Given the pleiotropic effect of global regulators, they must not belong to modules but rather coordinate them in response to general-interest environmental cues.
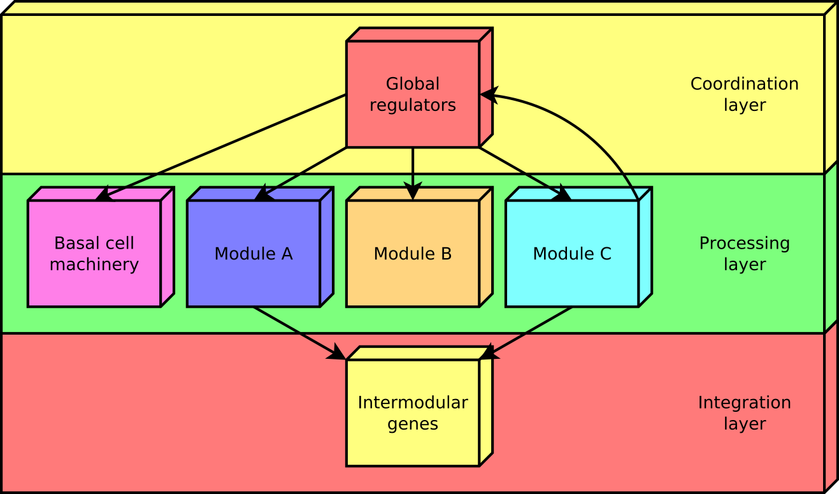
According to this approach, every gene in a regulatory network is predicted to belong to one out of four possible classes of systems-level elements, which interrelate in a non-pyramidal, three-tier, hierarchy shaping the functional architecture as follows: (1) Global regulators are responsible for coordinating both the (2) basal cell machinery, composed of exclusively globally regulated genes (EGRGs), and (3) locally autonomous modules (shaped by modular genes), whereas (4) intermodular genes integrate, at promoter level, physiologically disparate module responses eliciting a combinatorial processing of environmental cues (Figure 2). The latter a new element never before described in regulatory networks, which was first identified by the NDA.
There are a couple of advantages resulting from developing an ad-hoc method based on biological knowledge: (1) Global regulators (hubs) does not belong to any module. This is biologically important because they are not related to a particular physiologic function. (2) We identify that there are overlap among modules and it is mediated by the intermodular genes. None of this key features existing in bacterial regulatory networks could be identified with any of the methods commonly used to identify communities in complex networks. Finally, the NDA does not only provide a method to identify the system-level elements shaping bacterial regulatory networks, but it also paves the way towards a theory of the principles governing the organization and evolution of these cellular control networks.
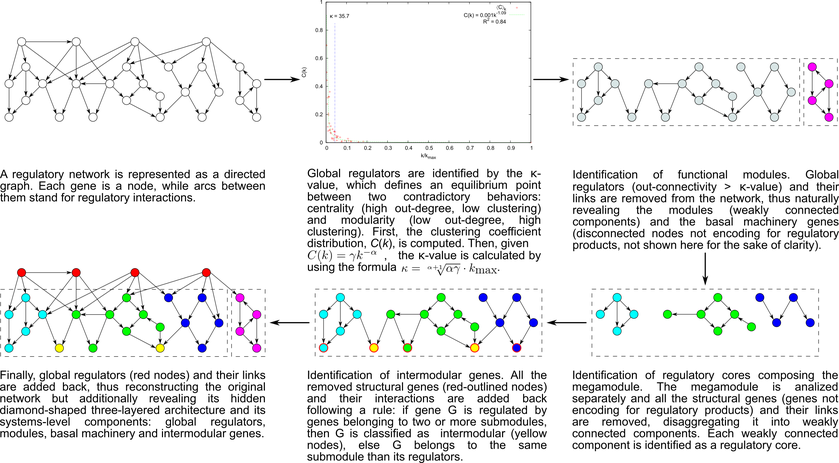
Figure 1. The natural decomposition approach.
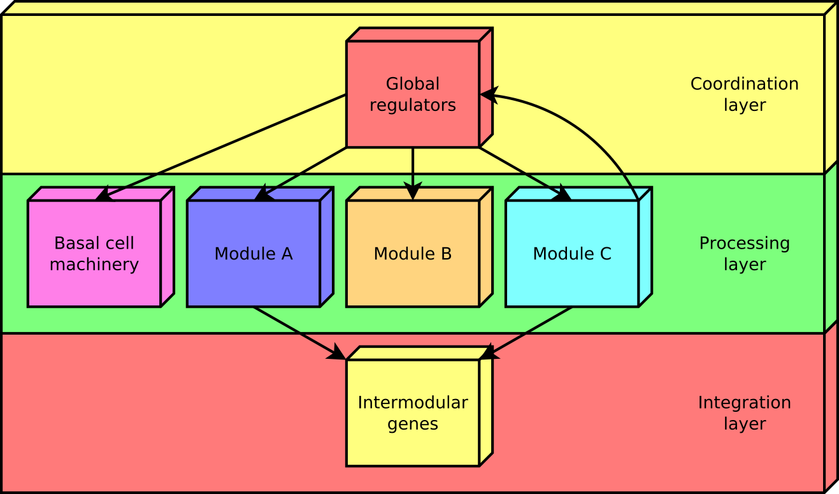
Figure 2. The functional architecture unveiled by the NDA is a diamond-shaped, three-tier hierarchy, exhibiting some feedback between processing and coordination layers, which is shaped by four classes of systems-level elements: global transcription factors, locally autonomous modules, basal machinery, and intermodular genes.
- Freyre-Gonzalez, J.A.*, Escorcia-Rodríguez, J.M., Gutiérrez-Mondragón, L.F., Martí-Vértiz, J. Torres-Franco C.N., Zorro-Aranda, A. System principles governing the organization, architecture, dynamics, and evolution of gene regulatory networks. Frontiers in Bioengineering and Biotechnology 10:888732 (2022) doi:10.3389/fbioe.2022.888732
- Escorcia-Rodríguez, J.M., Tauch, A., Freyre-González, J.A.* Corynebacterium glutamicum regulation beyond transcription: Organizing principles and reconstruction of an extended regulatory network incorporating regulations mediated by small RNA and protein-protein interactions. Microorganisms 9(7):1395 (2021) doi:10.3390/microorganisms9071395
- Escorcia-Rodríguez, J.M., Tauch, A., Freyre-González, J.A.* Abasy Atlas v2.2: The most comprehensive and up-to-date inventory of meta-curated, historical, bacterial regulatory networks, their completeness and system-level characterization. Computational and Structural Biotechnology Journal 18:1228-1237 (2020) doi:10.1016/j.csbj.2020.05.015
- Freyre-González, J.A.*, Tauch, A. Functional architecture and global properties of the Corynebacterium glutamicum regulatory network: novel insights from a dataset with a high genomic coverage. Journal of Biotechnology 257C:199-210 (2017) doi:10.1016/j.jbiotec.2016.10.025
- Ibarra-Arellano, M.A., Campos-González, A.I., Treviño-Quintanilla, L.G., Tauch, A., Freyre-González, J.A.* Abasy Atlas: A comprehensive inventory of systems, global network properties, and systems-level elements across bacteria. Database 2016:baw089 (2016) doi:10.1093/database/baw089
- Freyre-González, J.A.*, Treviño-Quintanilla, L.G., Valtierra-Gutiérrez, I., Gutierrez-Ríos, R.M., Alonso-Pavón, J.A. Prokaryotic regulatory systems biology: Common principles governing the functional architectures of Bacillus subtilis and Escherichia coli unveiled by the natural decomposition approach. Journal of Biotechnology 161(3):278-286 (2012) doi:10.1016/j.jbiotec.2012.03.028
- Freyre-Gonzalez, J.A.* and Trevino-Quintanilla, L.G. Analyzing regulatory networks in bacteria. Nature Education 3(9):24 (2010) http://www.nature.com/scitable/topicpage/analyzing-regulatory-networks-in-bacteria-14426192
- Freyre-González, J.A.*, Alonso-Pavón, J.A., Treviño-Quintanilla, L.G., and Collado-Vides, J. Functional architecture of Escherichia coli: new insights provided by a natural decomposition approach. Genome Biology 9(10):R154 (2008) doi:10.1186/gb-2008-9-10-r154